De Novo Design
·
Access
to >1020 molecules with molecular weight <500
·
Emphasis
on chemically stable and reasonable molecules
·
Space
searchable using any computable properties
·
User
can influence search towards more interesting hits, away from undesirable
moieties etc..
The
following examples are intended to show that the representation of chemistry
space produces reasonable molecules that match the desired criteria whether
that be fitting in a binding site of interest or similar to known ligands. None of these molecules have been synthesized
or tested.
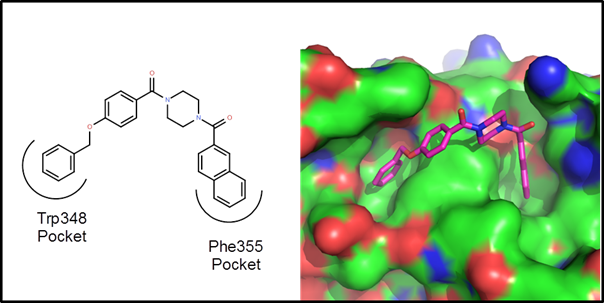
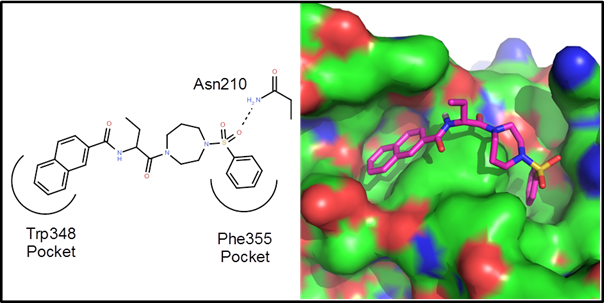
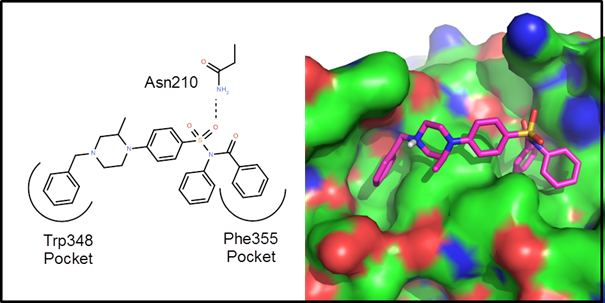
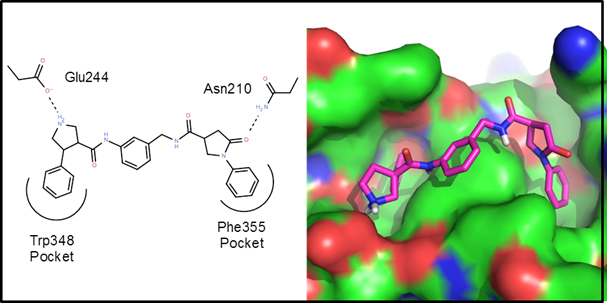
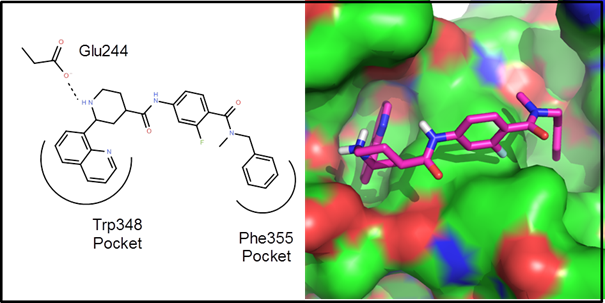
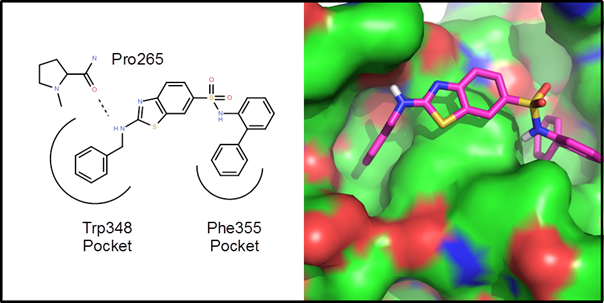
Example 1 – A protein-protein interaction. This is an example driven by docking/scoring
molecules in the binding site of the target protein. The protein structure, 2d10, is a co-crystal
structure of Radixin with a helical peptide from
NHERF1. NHERF1 is involved in CCR5
trafficking and as a result, could potentially be an HIV target. There are two rather deep pockets
corresponding to Trp345 and Phe355 of the NHERF1 helical peptide:

The goal of
this de novo design exercise was to find molecules that fit in these two
pockets and explore the variety of polar interactions in and around these
pockets. In all about 2 weeks of CPU time
(4 processors for 3.5 days) were used to generate, dock and score 25
generations of molecules. Below are 10 example
virtual hits: